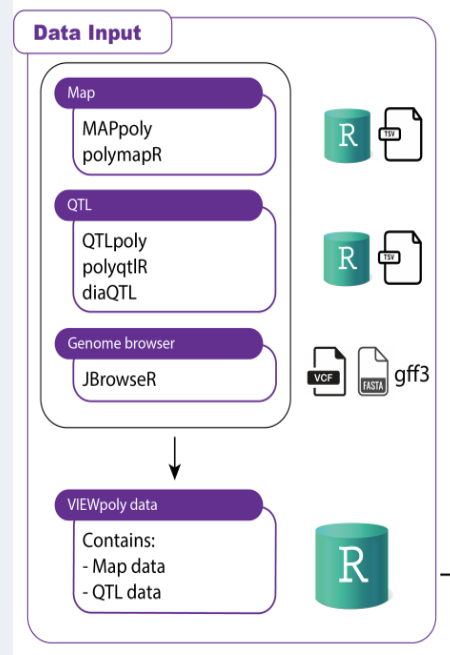
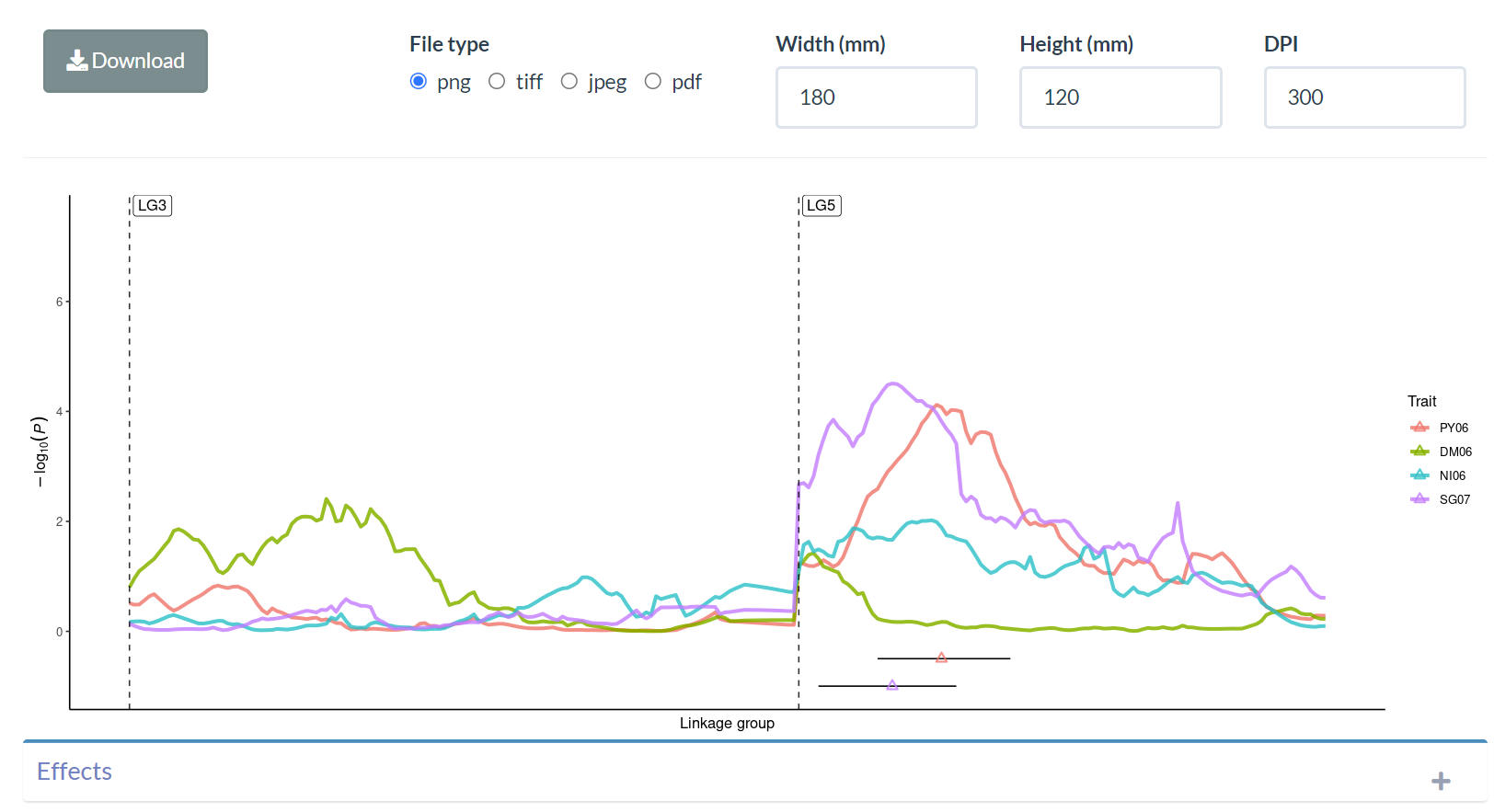
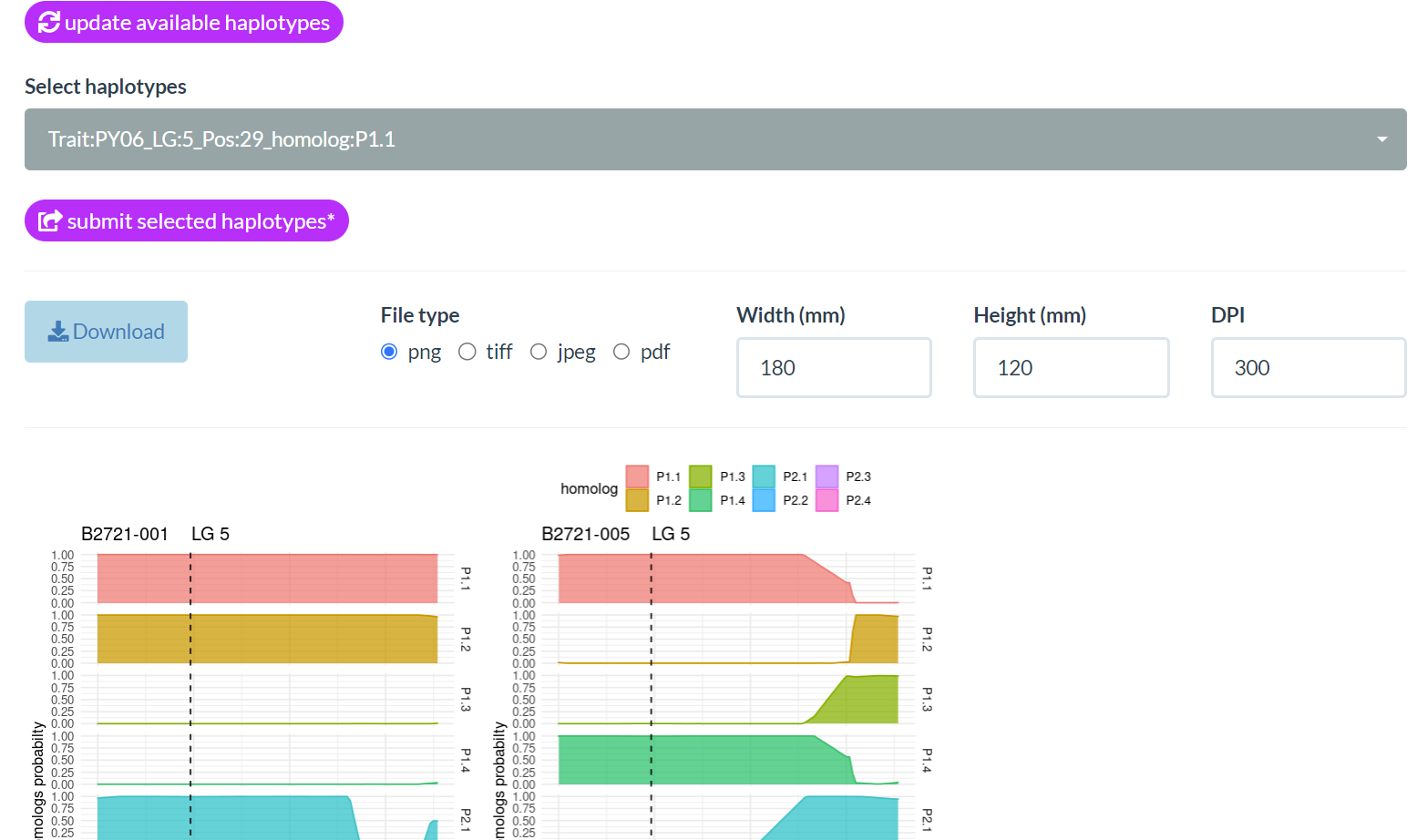
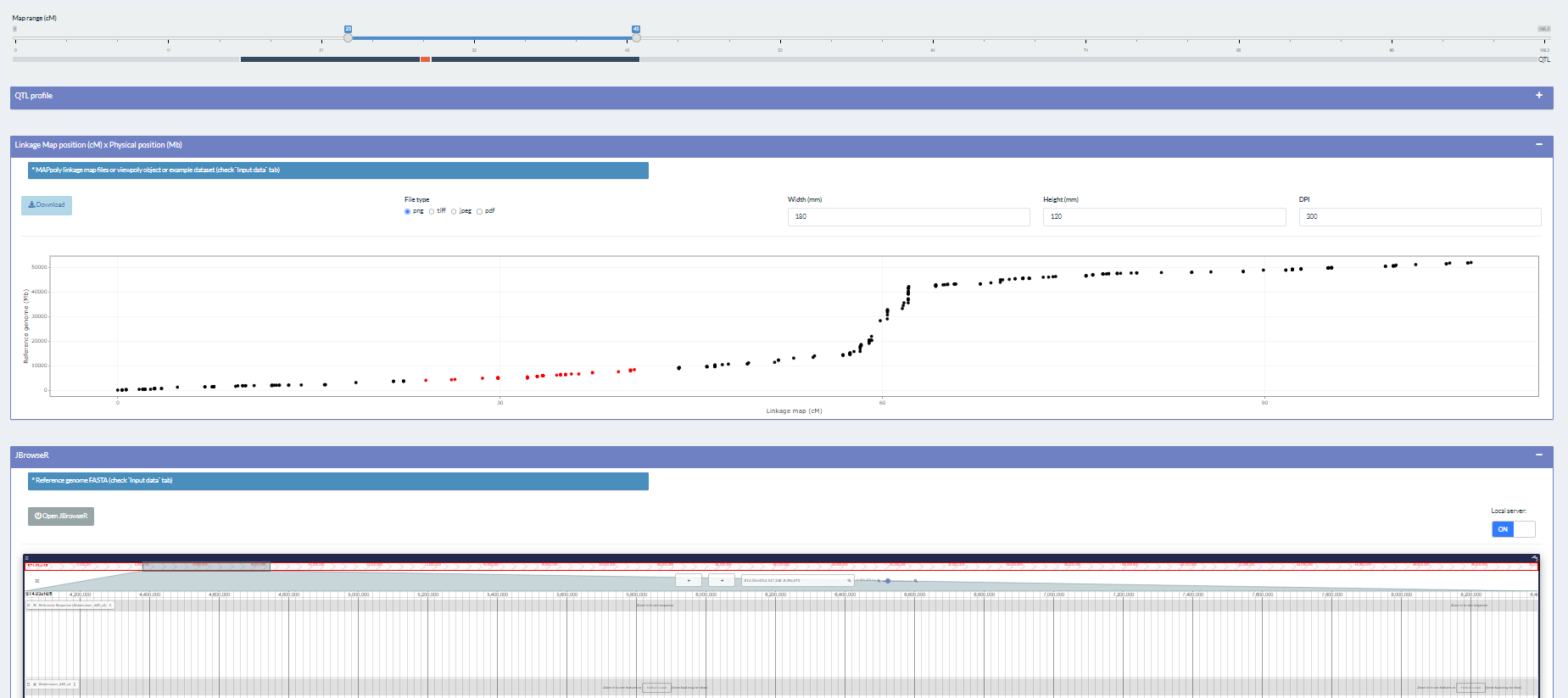
EXPERIENCE
Education
AWARDS
- USDA Secretary Honor Award 2024 - In a ceremony held on January 10, 2025, in Washington D.C., the U.S. Department of Agriculture (USDA) awarded Breeding Insight (BI) and BI OnRamp the prestigious Agricultural Secretary Honor Award in the category of “Providing all Americans safe, nutritious food”
- Postdoctoral Scholar Travel Award 2023 - Texas A&M University
- Market Ready Prize - Cornell Institute Digital Agriculture Hackathon 2021
- WikiProject Computational Biology/ISCB award 2018 - International Society for Computational Biology (ISCB) and WikiProject Computational Biology
- Best Poster Award - Brazilian Association for Bioinformatics and Computational Biology